
Research
DNA segregation in Caulobacter Crescentus
C. Crescentus uses active processes to segregate its DNA. The proteins ParB and ParA are though to be important in controlling segregation. Recent experiment point to similarities between the segregation apparatus in Crescentus and the mitotic spindle in higher eukaryotic cells. We have proposed a model for chromosome segregation based on the dynamic interactios between ParB binding sites and bundles of linear ParA filaments. I am am interested in modeling various aspects of chromosome segregation in bacteria such as generation of directed motion of chromosome copies and the biochemical control of chromosome movement directionality.B. Shtylla and J. P. Keener, A mathematical model of ParA filament-mediated chromosome motility in bacteria, Journal of Theoretical Biology, 2012, 307, pp. 82-95.
Polymer Brownian Motors
Biopolymer motors generate movement by using the energy of
polymerization/depolymerization. In contrast to molecular
motor enzymes such as dynein or kinesin, which use ATP and are
recruited and reused in various locations of the cell,
biopolymer motors are only assembled to perform work at
specific locations and are then dissasembled. In mammalian
cells, chromosomes interact with many dynamic microtubules
while they move to the cell equator. We have proposed a
mathematical model for how kinetochore filaments can be
coupled with dynamic microtubules to generate movement. We
find that the resulting force-velocity relations are
non-linear and that motility characteristics depend on the
individual kinetochore filament binding strengths. I am
interested in modeling the dynamic interactions between
various kinetochore proteins and microtubules. In general, I
am interested in the collective behavior of active and passive
molecular motor assays. The details of interactions between
these cellular machines are important for the formation of
eukaryotic mitotic spindles and the proper progression of cell
division.

J. P. Keener and B. Shtylla. A mathematical model of force generation by flexible kinetochore-microtubule attachments*, Biophysical Journal, 106 (5), pp. 998-1007. abstract
*Article featured on the cover of the March 04 issue of the Biophysical Journal and in "Modeling kinetochores: when form and function becomes art" on the Biophysical Society Blog
A. Sharma*, B. Shtylla* and D. Chowdhury. Distribution of lifetimes of kinetochore-microtubule attachments:interplay of energy landscape, molecular motors and microtubule (de-)polymerization, 2013, (to appear in Physical Biology).
B. Shtylla and J. P. Keener. A mathematical model for force generation at the kinetochore-microtubule interface. SIAM Journal on Applied Mathematics, 2011, 71(5), pp. 1821-1848. abstract, pdf
Mechano-chemical control of Mitotic events in higher
Eukaryotes
During mitosis, cells have to ensure that precisely half of
the replicated DNA is allocated to each daughter cell.
Accuracy is achieved with the help of a complex biochemical
regulatory network, which keeps track of the microtubule
attachment status of each moving chromosome during division.
Even a single lost attachment will put division into a hold!
The surveillance of a chromosome requires that a cell know not
only the attachment status but also the amount of force that
each chromosome is sustaining due to its interactions with the
mitotic spindle. We have shown that mechanochemical feedbacks
are important in assuring proper mitosis progression. I am
interested in modeling the mechanisms mediating the
integration of biochemical and mechanical signals during
mitosis in mammalian cells.
Publications:
A. Matzavinos, B. Shtylla, Z. Voller, S. Liu, and M. A.J. Chaplain. Stochastic modelling of chromosomal segregation: Errors can introduce correction, 2013, (in revision).
Full list of publications
Undergraduate Research Opportunities:
There are various venues for participation in research for students that are interested in mathematical biology:
1. Research assistant (academic year or summer): contact me directly if interested.
2. 5C SURP or HHMI funded fellows can work on problems in mathematical biology with me here.
3. Senior Thesis in Mathematics as part of the senior exercise, if you are a double major looking for a primer on interdisciplinary research this might be a great option. It is preferable that you contact me during your Junior year to explore options. Some generic instructions about senior thesis in mathematics are here and for interdisciplinary projects here. I am happy to discuss Mathematics Thesis research projects, especially if you are a junior math major at Pomona college. Some examples of thesis projects I have supervised are listed below.
Senior Thesis:
2013-2014
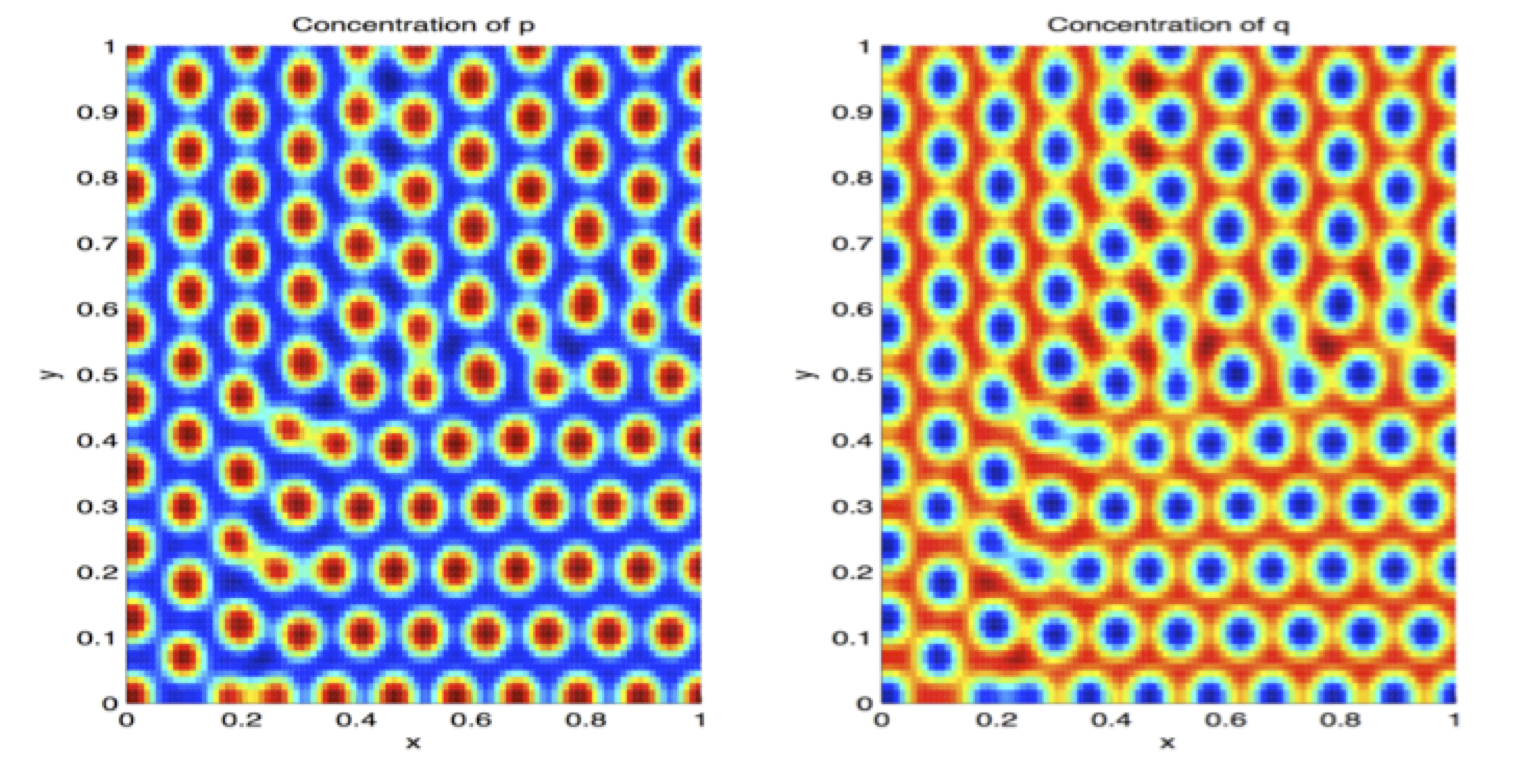
- Emily Meyer, Pomona '14: Turing Patterns for spatial-temporal signalling. Emily is working on reaction-diffusion models for protein localization in dividing cells.
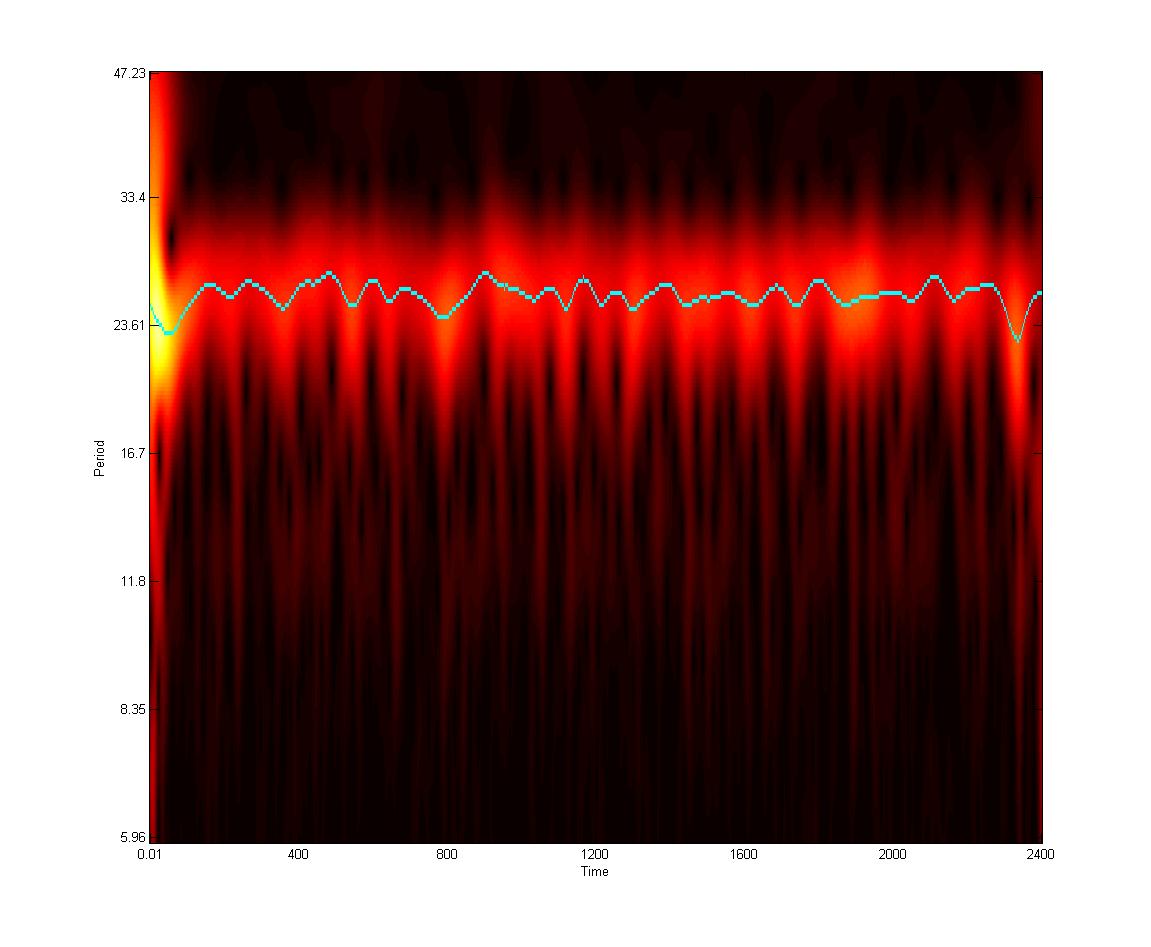
- David Morgens, Pomona '14. Noisy Clocks: Stochastic Modeling of Circadian Rhythms. David is working on analysis of stochastic fluctuations in order to identify functional biological modules.
- Kyle Roskamp, Pomona '14. Understanding and Applying Graph Ranking to Gene Ontologies.
- Kevin Guttenplan, Pomona '14. Mathematical Modeling of Axon Guidance at the Drosophila melanogaster Midline.
A full schedule of our exciting thesis presentations and topics can be found here . If you want to find out more about this, come and join us for the talks!
Current Research Students:
- Matt McDermott, Havery Mudd College '14: A computational model for cell mechanics in early C. elegans development.
Previous Research Students:
- Sangeeta Warrier, Mt. Holyoke College '15.
Current Research Funding:
National Science Foundation RUI (2012-2015)
"Mathematical Modeling of chromosome segregation and organization in bacteria"