Origins and Evolution of the genetic code:
Introduction:
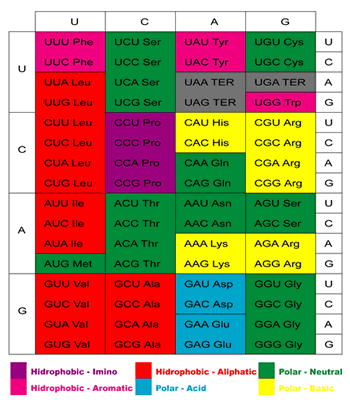
The genetic code (Figure 1) is the set of rules that defines the correspondence between the 20 amino acids in proteins and groups of three bases – codons – in the mRNAs.
There are several theories that try to explain the origin of the code. Most can be classified in one of three major groups (Knight et al. 1999; Cavalcanti and Landweber 2004 is a short review of upstanding questions regarding the genetic code).
-
Chemical: Posits that direct chemical interactions between amino acids and their cognate codons/anticodons
influenced codon assignment. Studies of binding of RNA aptamers to amino acids showed that, for at least some amino acids
– Arginine, Tyrosine and Isoleucine – such chemical interactions do in fact exist. These theories fail to
explain the assignment of codons that do not show direct interactions to their cognate amino acids.
-
Historical: Proposes that an initially smaller code grew by incorporation of new amino acids. For example, new amino acids
may have captured codons from their metabolic precursors, contributing to the assignment of similar amino acids to similar
codons.
-
Selection: suggests that the code was selected to minimize the phenotypic effects of point mutations. The code’s organization
supports this: nonsynonymous substitutions often lead to replacement of an amino acid by one chemically similar, causing
little disruption in the protein. Because of this tendency for point mutations to cause little changes in the amino acids, the
code is said to be minimized with respect to point mutations.
Accumulation of supporting evidence for each of these models suggests that they are not mutually exclusive. Rather, the code probably evolved by an interplay among some or all of them. Direct interactions of short RNA molecules and amino acids may have fixed the assignment of certain codons, while subsequent assignments may have been driven by history and selection.
The Aminoacyl-tRNA Synthetases and the Genetic Code:
The aminoacyl-tRNA synthetases – aaRS – are the enzymes that charge the tRNAs with their cognate amino acids. The aaRS can be divided in two classes, Class I – which contains the enzymes responsible for charging the tRNAs cognate to Arg, Cys, Gln, Glu, Ile, Leu, Met, Trp, Tyr ald Val – and Class II – Ala, Asn, Asp, Gly, His, Lys, Phe, Pro, Ser and Thr. These two enzyme classes are supposed to have originated independently.
It was long thought that these enzymes played an important role in shapping the genetic code, and my research focused in trying to understand this role. In Cavalcanti et al (2000) we showed that the observed minimization of the code with regard to point mutations is due mostly to point mutations that do not change the class of the amino acids coded. These mutations are highly minimized, while the mutations that do change the class are only moderately minimized.
Recently, we showed (Cavalcanti et al 2004) that the division of the aaRS in two classes is highly correlated with the properties of the amino acids in each class. Furthermore, the evolutionary tree of the enzymes in each class is very similar to dendrograms for the amino acids in that class, built using a several amino acid parameters from the aaindex database (Kawashima and Kanehisa 2000).
Evolution of the genetic code:
Although most organisms have the same genetic code, researchers began to discover exceptions to the ‘universal’ code in 1979, and today we know of more than 15 alternative codes (see a list at NCBI); each has just a few differences from the standard code, indicating common ancestry from this code. Several of these codes arose independently a number of times in evolution and are present in a variety of taxa.
How and why some organisms re-assign some of their codons, and the evolutionary consequences of such re-assignments are still hotly debated (Knight et al 2001). Currently the genomes of several eukaryotes that re-assign stop codons are being sequenced – Tetrahymena termophila ( http://www.ciliate.org/), Paramecium tetraurelia (http://paramecium.cgm.cnrs-gif.fr/), Oxytricha trifallax (http://oxytricha.princeton.edu/genome/, Doak et al 2003; Cavalcanti et al 2004a, b). We hope to use these genome sequences to shed some light in these questions.
Cavalcanti A.R.O., Neto B.B., Ferreira R. (2000) On the Classes of Aminoacyl-tRNA Synthetases and the Error Minimization in the Genetic Code., J.Theor.Biol.204, 15-20. ![]()
Cavalcanti A.R.O., Leite E.S., Neto B.B., Ferreira R. (2004) Correlation of the Classes of aaRS and their Cognate Amino Acids. Orig. Life Evol. Biosph.34 (4): 407-420.
Cavalcanti A.R.O., Landweber L.F. (2004) Quick Guide: Genetic Code. Curr. Biol.14 (4): R147. ![]()
Cavalcanti A.R.O., Dunn D.M., Weiss R., Herrick G., Landweber L.F., Doak T.G. (2004a) Sequence features of Oxytricha trifallax (class Spirotrichea) macronuclear telomeric and subtelomeric sequences. Protist (in press).
Cavalcanti A.R.O., Stover N., Orecchia L., Doak T.G., Landweber L.F. (2004b) Coding properties of Oxytricha trifallax (Sterkiella histriomuscorum) macronuclear chromosomes: analysis of a pilot genome project. Chomosoma (in press).
Doak T.G., Cavalcanti A.R.O., Stover N., Dunn D.M., Weiss R., Herrick G., Landweber L.F. (2003) Sequencing the Oxytricha trifallax macronuclear genome: a pilot project. Trends Genet. 19 (11): 603-607. ![]()
Kawashima S., Kanehisa M. (2000) AAindex: Amino acid index database. Nucleic Acids Res 28: 374. ![]()
Knight R.D., Freeland S.J., Landweber L.F. (1999) Selection, history and chemistry: The three faces of the genetic code. Trends Biochem Sci 24 (6): 241-247. ![]()
Knight R.D., Freeland S.J., Landweber L.F. (2001). Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet 2, 49–58. ![]()