This web page was produced as an assignment for a course on
Statistical Analysis on Microarray Data at
In this section I discuss running Significance Analysis of Microarrays (SAM) on the microarray data for this project.
From this section and on, I have combined spots into single miRNA by taking the average of the M value for the two spots that go together (if both spots are good, or just the good spot if one spot is bad, or NA if both spots are bad).
Because there was both tumor tissue and normal tissue used in the study, the first SAM that I ran was comparing the miRNA of the tumor and normal tissue.
SAM is testing the hypothesis for each miRNA (i.e. 768 times hypothesis tests since there are 768 distinct miRNA per array)
H0: the miRNA is not able to discriminate between tumor tissue and normal tissue
Ha0: the miRNA is able to discriminate between tumor tissue and normal tissue
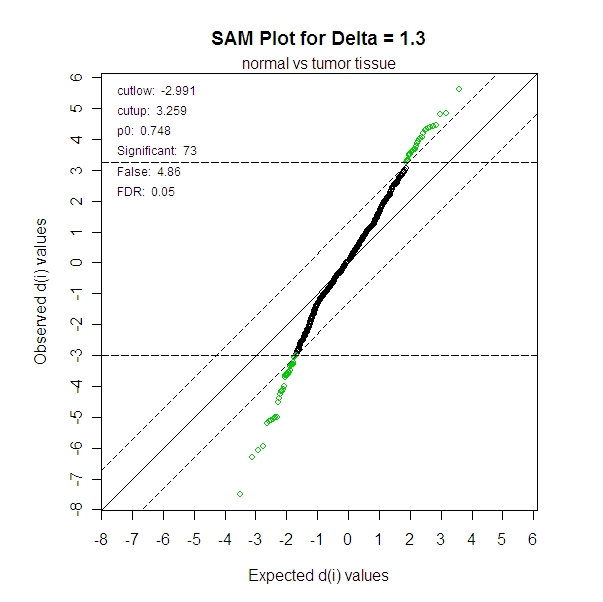
With a False Discovery Rate (FDR) set at 0.05, there were 73 miRNA that were found to be significant. In other words, when we set the expect to see about 1 in 20 falsely reported significant genes in the whole experiment, then there were 73 miRNA for which a t-test rejected the null hypothesis.
>
summary(norm.sam.tumor,1.3)
SAM
Analysis for the Two-Class Unpaired Case Assuming Unequal Variances
s0
= 0
Number of permutations: 100
MEAN number of falsely called variables is
computed.
Delta:
1.3
cutlow:
-2.991
cutup:
3.259
p0:
0.748
Identified
Genes: 73
Falsely
Called Genes: 4.86
FDR:
0.0498
Identified
Genes (using Delta = 1.3):
Row d.value
stdev rawp q.value R.fold Name
1 429 -7.49 0.0802 0
0 0.6676
2 50 -6.29 0.1798 1.30e-05 0.00187 0.4682
hsa_miR_431
3 553 -6.08 0.1005 1.30e-05 0.00187 0.6522
CONTROL_2
4 124 -5.96 0.2319 1.30e-05 0.00187 0.3823
CONTROL_1
5 129 5.60 0.3399 6.51e-05 0.00686 3.4471
hsa_miR_135b
6 394 -5.19 0.2599 0.000117 0.00686 0.4204
hsa_miR_513
7 317 -5.13 0.2517 0.000117 0.00686 0.4210
8 688 -5.11 0.7208 0.000117 0.00686 0.0972
hsa_miR_196a
9 167 -5.08 0.1655 0.000117 0.00686 0.5558
ambi_miR_2537
10 70 -5.04 0.3538 0.000130 0.00686 0.3037
11 724 -5.00
0.1645 0.000143 0.00686 0.6032
12 263 -5.00
0.0793 0.000143 0.00686 0.7661
13 567 4.84
0.5783 0.000169 0.00695 11.5361
hsa_miR_151
14 221 4.81
0.4253 0.000169 0.00695 4.1530 hsa_miR_214
15 664 -4.52
0.0588 0.000365 0.01171 0.8352 ambi_miR_7081
16 759 4.44
0.7369 0.000391 0.01171 8.4939 hsa_miR_126
17 314 4.42
0.2185 0.000391 0.01171 2.0808 hsa_miR_150
18 602 4.40
0.1699 0.000404 0.01171 1.9313
19 703 -4.39
0.1984 0.000404 0.01171 0.6587
20 583 4.37
0.1156 0.000443 0.01171 1.4630 hsa_miR_143
21 530 4.32
0.2654 0.000456 0.01171 2.0899 CONTROL_8
22 82 4.30
0.1891 0.000456 0.01171 1.8157 CONTROL_1
23 45 -4.28 0.1874 0.000469 0.01171 0.6019
24 643 4.19
0.1087 0.000651 0.01372 1.3868 ambi_miR_4482
25 43 -4.17 0.2719 0.000651 0.01372 0.5757
26 141 -4.17
0.1551 0.000651 0.01372 0.6311 ambi_miR_10594
27 667 -4.16
1.0041 0.000651 0.01372 0.0651 ambi_miR_4482
28 26 -4.10 0.2103 0.000703 0.01372 0.5976
hsa_miR_125b
29 247 4.08
0.1909 0.000703 0.01372 1.6710 hsa_miR_130a
30 680 4.04 0.1223
0.000716 0.01372 1.4122 hsa_miR_329
31 509 4.01
0.6301 0.000755 0.01400 3.1442 hsa_miR_22
32 380 -4.00
0.2463 0.000794 0.01426 0.5555 ambi_miR_10202
33 6 3.94 0.3079 0.000872 0.01519 3.2538
hsa_miR_181b
34 415 3.86
0.1400 0.001133 0.01915 1.4606
hsa_miR_302c_AS
35 570 3.78
1.4356 0.001380 0.02266 3.8711 hsa_miR_345
36 259 -3.72
0.9339 0.001628 0.02598 0.0590 ambi_miR_11576
37 218 3.68
0.2444 0.001745 0.02658 1.8184 hsa_miR_181c
38 4 3.67 0.1303 0.001758 0.02658 1.4380
39 88 -3.65 0.2594 0.001888 0.02668 0.5196
hsa_miR_365
40 375 -3.64
0.3411 0.001914 0.02668 0.5682 hsa_miR_141
41 318 -3.64
0.5404 0.001927 0.02668 0.2563 hsa_miR_511
42
206 3.63 0.1848 0.002044 0.02668 1.9137
43
702 -3.62 0.5295 0.002083 0.02668 0.2248
hsa_miR_346
44 644 3.61
0.1150 0.002135 0.02668 1.3464 ambi_miR_12180
45 515 -3.61
0.2264 0.002135 0.02668 0.5403
46 320 3.61
0.2805 0.002135 0.02668 2.6407 hsa_miR_299_3p
47 416 3.57
0.1961 0.002357 0.02825 1.7136 hsa_miR_199b
48 316 -3.56
0.1843 0.002370 0.02825 0.6219 hsa_miR_18a_AS
49 32 -3.54 0.2444 0.002409 0.02825 0.6840
hsa_miR_486
50 587 -3.54 0.4721 0.002526 0.02903 0.3323
51 123 3.52
0.1632 0.002669 0.03008 1.4475 hsa_let_7a
52 555 3.50
0.6640 0.002826 0.03123 3.3120 CONTROL_27
53 646 -3.48
0.1084 0.002969 0.03215 0.7856
54 289 3.47
0.3999 0.003021 0.03215 2.5268 hsa_miR_335
55 654 -3.36
0.6197 0.003828 0.04000 0.2992 hsa_miR_424
56 147 3.33
0.1170 0.004063 0.04062 1.3295 CONTROL_23
57 199 3.32
0.1121 0.004154 0.04062 1.3162 hsa_miR_519c
58 502 -3.31 0.2809
0.004284 0.04062 0.5542 ambi_miR_2825
59 121 -3.31
0.6430 0.004284 0.04062 0.2520
60 489 -3.30
1.2215 0.004297 0.04062 0.0507
61 35 -3.29 0.3712 0.004375 0.04062 0.5171
hsa_miR_34b
62 581 3.29
0.1322 0.004388 0.04062 1.3492 hsa_miR_383
63 674 -3.28
0.1752 0.004453 0.04062 0.6944 hsa_miR_412
64 310 3.26
0.1570 0.004661 0.04186 1.6397
65 656 -3.22
0.0578 0.005065 0.04444 0.8813 hsa_miR_210
66 562 -3.21
0.3542 0.005104 0.04444 0.4645 hsa_miR_30a_3p
67 241 -3.07
0.4398 0.006680 0.05729 0.4082 hsa_miR_222
68 706 -3.05
0.4624 0.006901 0.05748 0.3804
69 540 -3.05
0.4133 0.007031 0.05772 0.6832 hsa_miR_103
70 162 -3.03
0.2049 0.007370 0.05936 0.6998 hsa_miR_149
71
383 -3.02 0.0910 0.007448 0.05936 0.8346
ambi_miR_3121
72
113 -3.01 0.4564 0.007552 0.05936 0.3554
73
295 -2.99 1.2154 0.007930 0.05996 0.0929
In this SAM, each miRNA is testing
H0: All the tissue types are the same
Ha: The tissue types are not all the same
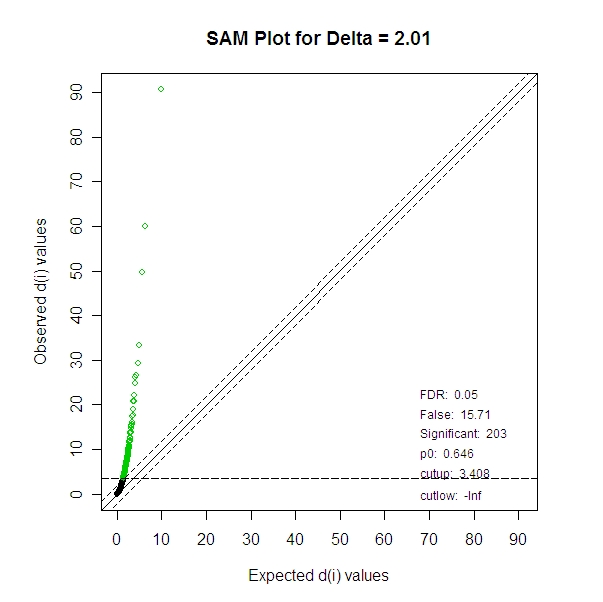
With a FDR of 0.05, there were 203 miRNA identified as significant.
>
summary(norm.sam.all,2.01)
SAM
Analysis for the Multi-Class Case with 7 Classes
s0
= 0
Number of permutations: 100
MEAN number of falsely called variables is
computed.
Delta:
2.01
cutlow:
-Inf
cutup:
3.408
p0:
0.646
Identified
Genes: 203
Falsely
Called Genes: 15.71
FDR:
0.05
Identified
Genes (using Delta = 2.01):
Row d.value
stdev rawp q.value R.fold Name
1 570 90.62 0.4510 1.30e-05 0.00323 NA
hsa_miR_345
2 509 59.98 0.1392 1.30e-05 0.00323 NA
hsa_miR_22
3 561
49.65 0.6024 2.60e-05 0.00430
NA
4 185 33.31 1.2372 5.21e-05 0.00549 NA
hsa_miR_202
5 27 29.26 0.8496 7.81e-05 0.00549 NA
hsa_miR_337
6 259
26.54 0.7181 7.81e-05 0.00549
NA ambi_miR_11576
7 489
26.12 1.3981 9.11e-05 0.00549
NA
8 295
24.78 1.5405 0.000104 0.00549
NA
9 699
22.21 1.8141 0.000143 0.00549
NA
10 101 20.81 0.7893 0.000182 0.00549 NA
hsa_miR_380_5p
11 755 20.75 1.4384 0.000182 0.00549 NA
12 605 18.87 0.0402 0.000208 0.00549 NA
hsa_miR_383
13 8 17.74 0.4164 0.000234 0.00549 NA
hsa_miR_486
14 452 17.39 0.4578 0.000247 0.00549 NA
ambi_miR_10766
15 728
15.99 0.3031 0.000286 0.00549
NA
16 555 15.70 0.5351 0.000286 0.00549 NA
CONTROL_27
17 688 15.42 1.3906 0.000286 0.00549 NA
hsa_miR_196a
18 359
15.22 1.0307 0.000286 0.00549
NA ambi_miR_3121
19 281 15.14 0.5524 0.000299 0.00549 NA
ambi_miR_13237
20 377 14.78 1.0609 0.000299 0.00549 NA
hsa_miR_133b
21 123
14.74 0.0282 0.000299 0.00549
NA hsa_let_7a
22 678 13.83 1.4517 0.000352 0.00549 NA
hsa_miR_346
23 609 13.73 0.6230 0.000352 0.00549 NA
hsa_let_7d
24 667
13.54 2.0535 0.000365 0.00549
NA ambi_miR_4482
25 298
12.68 0.6557 0.000378 0.00549
NA
26 467
12.53 0.0770 0.000391 0.00549
NA hsa_miR_326
27 121
12.04 0.8750 0.000404 0.00549
NA
28 536 11.83 0.5268 0.000404 0.00549 NA
hsa_miR_503
29 141 11.80 0.0496 0.000404 0.00549 NA
ambi_miR_10594
30 665
11.74 0.9858 0.000404 0.00549
NA ambi_miR_13090
31 474 11.57 2.1651 0.000404 0.00549 NA
ambi_miR_13266
32 701 11.49 0.3756 0.000404 0.00549 NA
hsa_miR_523
33 187
10.98 0.4445 0.000417 0.00549
NA ambi_miR_7075
34 337 10.80 1.5958 0.000430 0.00549 NA
CONTROL_30
35 602 10.63 0.2003 0.000430 0.00549 NA
36 379 10.57 0.3257 0.000430 0.00549 NA
ambi_miR_3998
37 515 10.52 0.0846 0.000430 0.00549 NA
38 255 10.38 1.0417 0.000443 0.00549 NA
39 734 10.27 1.3090 0.000443 0.00549 NA
hsa_miR_485_5p
40 719 10.20 0.1140 0.000443 0.00549 NA
41 528 10.03 0.1256 0.000456 0.00551 NA
ambi_miR_9534
42 198 9.81 0.9123 0.000469 0.00553 NA
hsa_miR_522
43 694
9.67 0.0838 0.000495 0.00558
NA
44 61
9.54 0.1297 0.000495 0.00558
NA
45 214
9.54 0.6210 0.000508 0.00559
NA
46 274 9.02 0.1499 0.000573 0.00580 NA
47 207 9.00 0.1860 0.000573 0.00580 NA
hsa_miR_340
48 158 8.92 0.3045 0.000573 0.00580 NA
hsa_miR_134
49 247 8.87 0.0924 0.000573 0.00580 NA
hsa_miR_130a
50 35 8.71 0.5420 0.000586 0.00581 NA
hsa_miR_34b
51 162
8.64 0.1714 0.000612 0.00595
NA hsa_miR_149
52 241 8.38 0.5268 0.000677 0.00633 NA
hsa_miR_222
53 387 8.37 0.4565 0.000677 0.00633 NA
hsa_miR_9_AS
54 65
8.15 0.2991 0.000729 0.00669
NA ambi_miR_7084
55 306 7.96 1.5294 0.000820 0.00739 NA
hsa_miR_410
56 129 7.78 0.4497 0.000885 0.00744 NA
hsa_miR_135b
57 553
7.77 0.0482 0.000885 0.00744
NA CONTROL_2
58 223 7.76 0.0862 0.000885 0.00744 NA
hsa_miR_519c
59 106 7.73 0.4998 0.000898 0.00744 NA
60 522 7.72 0.2124 0.000911 0.00744 NA
61 370 7.61 1.1009 0.000951 0.00744 NA
hsa_miR_128a
62 676 7.50 0.1863 0.000990 0.00744 NA hsa_miR_518f_AS
63 586
7.47 0.3822 0.001003 0.00744
NA
64 626
7.39 0.0585 0.001029 0.00744
NA
65 91
7.37 0.1455 0.001055 0.00744
NA ambi_miR_9630
66 86
7.36 0.1286 0.001055 0.00744
NA
67 560 7.32 0.5223 0.001068 0.00744 NA
hsa_miR_503
68 742 7.28 0.4407 0.001081 0.00744 NA
ambi_miR_4209
69 643
7.27 0.0555 0.001081 0.00744
NA ambi_miR_4482
70 316 7.24 0.0918 0.001081 0.00744 NA
hsa_miR_18a_AS
71 680 7.24 0.0577 0.001081 0.00744 NA
hsa_miR_329
72 513
7.18 1.0381 0.001081 0.00744
NA
73 432 7.14 0.7236 0.001120 0.00760 NA
74 291 6.77 1.0119 0.001380 0.00925 NA
ambi_miR_13124
75 629 6.64 1.2765 0.001432 0.00934 NA
hsa_miR_517a
76 178 6.64 0.5254 0.001432 0.00934 NA
hsa_miR_296
77 5 6.50 0.2330 0.001549 0.00957 NA
hsa_miR_375
78 624
6.49 0.3552 0.001549 0.00957
NA
79 124 6.48 0.3145 0.001563 0.00957 NA
CONTROL_1
80 318 6.44 0.9577 0.001589 0.00957 NA
hsa_miR_511
81 125
6.44 0.2093 0.001602 0.00957
NA hsa_miR_380_5p
82 225 6.40 0.1823 0.001602 0.00957 NA
hsa_miR_384
83 330 6.39 0.3864 0.001602 0.00957 NA
hsa_miR_410
84 245
6.32 0.7378 0.001667 0.00984
NA
85 446 6.28 1.0006 0.001719 0.00991 NA
86 627 6.28 0.8585 0.001719 0.00991 NA
hsa_miR_10b
87 762 6.25 0.5156 0.001758 0.01000 NA
hsa_miR_155
88 657 6.21 1.4104 0.001810 0.01000 NA
hsa_miR_377
89 53 6.20 0.3791 0.001810 0.01000 NA
hsa_miR_324_5p
90 642
6.19 0.8817 0.001823 0.01000
NA ambi_miR_13190
91 417 6.18 0.6318 0.001849 0.01000 NA
hsa_miR_7
92 705 6.16 0.3942 0.001862 0.01000 NA
hsa_miR_186
93 314
6.16 0.2751 0.001875 0.01000
NA hsa_miR_150
94 385 6.11 0.1642 0.001927 0.01016 NA
hsa_miR_512_3p
95 588 6.07 0.5137 0.001992 0.01040 NA
96 6 6.02 0.7116 0.002018 0.01042 NA
hsa_miR_181b
97 551
5.78 0.3645 0.002357 0.01205
NA ambi_miR_10394
98 724 5.69 0.2075 0.002461 0.01245 NA
99 279
5.67 0.0650 0.002487 0.01245
NA
100 380 5.64
0.3623 0.002526 0.01246 NA ambi_miR_10202
101 360 5.63
0.4384 0.002539 0.01246 NA ambi_miR_10133
102
702 5.62 0.9309 0.002565 0.01247 NA
hsa_miR_346
103 4 5.57 0.0767 0.002656 0.01279 NA
104 277 5.43
0.3920 0.002969 0.01415 NA
105 616 5.42
0.3573 0.003021 0.01425 NA ambi_miR_13209
106 50 5.41 0.2707 0.003047 0.01425 NA
hsa_miR_431
107 541 5.31
0.5101 0.003216 0.01475 NA hsa_miR_495
108
167 5.31 0.1351 0.003216 0.01475 NA
ambi_miR_2537
109 438 5.31
0.3974 0.003242 0.01475 NA
110 248 5.25
0.1061 0.003411 0.01529 NA hsa_miR_425
111 49 5.24 0.0515 0.003424 0.01529 NA
hsa_miR_373_AS
112 323 5.22
1.0414 0.003503 0.01550 NA
113 335 5.17
0.0980 0.003620 0.01574 NA
114 601 5.17
0.3991 0.003620 0.01574 NA hsa_miR_132
115 689 5.15
0.1253 0.003698 0.01586 NA hsa_miR_520b
116 608 5.14
0.3164 0.003711 0.01586 NA hsa_miR_525
117
600 5.12 1.0733 0.003776 0.01600 NA
118 317 5.07
0.3894 0.003854 0.01619 NA
119 64 5.00 0.3744 0.004010 0.01671 NA
hsa_miR_365
120 765 4.96
0.0514 0.004193 0.01729 NA ambi_miR_7920
121 753 4.96
0.3925 0.004219 0.01729 NA
122 415 4.87
0.0870 0.004583 0.01853 NA
hsa_miR_302c_AS
123 23
4.85 0.3127 0.004596 0.01853
NA
124 289 4.82
0.5283 0.004687 0.01874 NA hsa_miR_335
125 698 4.81
0.1875 0.004753 0.01885 NA hsa_miR_412
126
530 4.78 0.2495 0.004857 0.01897 NA
CONTROL_8
127 514 4.77
0.6560 0.004909 0.01897 NA
128 168 4.77
0.3726 0.004909 0.01897 NA ambi_miR_5
129 409 4.76
0.4048 0.004935 0.01897 NA hsa_miR_512_3p
130 766 4.75
0.1348 0.005013 0.01897 NA ambi_miR_4209
131 56 4.74 0.8029 0.005052 0.01897 NA
hsa_miR_26a
132
256 4.74 0.1959 0.005052 0.01897 NA
ambi_miR_7103
133 633 4.72
0.3981 0.005130 0.01912 NA hsa_miR_377
134 113 4.66
0.6786 0.005469 0.02023 NA
135 349 4.63
0.6822 0.005651 0.02075 NA hsa_miR_517c
136 707 4.59
0.3948 0.005820 0.02122 NA
137 628 4.57
0.6183 0.005924 0.02144 NA hsa_miR_505
138 2 4.56 1.3474 0.006029 0.02166 NA
hsa_miR_125b
139 145 4.53
1.3327 0.006341 0.02262 NA CONTROL_1
140 218 4.51
0.2094 0.006406 0.02269 NA hsa_miR_181c
141
683 4.49 1.2486 0.006523 0.02294 NA
142 391 4.47
0.7266 0.006641 0.02302 NA
hsa_miR_302c_AS
143 437 4.47
0.5881 0.006641 0.02302 NA hsa_miR_148a
144 28
4.45 0.0714 0.006732 0.02308
NA
145
512 4.44 0.2384 0.006771 0.02308 NA
146
429 4.43 0.0812 0.006797 0.02308 NA
147
132 4.39 0.3414 0.006979 0.02347 NA
148
130 4.38 0.7052 0.007005 0.02347 NA
149 764 4.37
0.5077 0.007083 0.02357 NA ambi_miR_8488
150 315 4.35
2.4541 0.007188 0.02376 NA ambi_miR_13124
151 527 4.31
0.1370 0.007461 0.02450 NA ambi_miR_5074
152 414 4.30
2.4187 0.007539 0.02459 NA hsa_miR_221
153 579 4.29
0.4936 0.007708 0.02498 NA hsa_miR_100
154 411 4.27
0.6346 0.007878 0.02532 NA hsa_miR_9_AS
155 394 4.26
0.6050 0.007917 0.02532 NA hsa_miR_513
156 221 4.24
1.0840 0.008073 0.02566 NA hsa_miR_214
157 640 4.20
1.4436 0.008490 0.02681 NA ambi_miR_7081
158 144 4.17
0.2970 0.008737 0.02741 NA ambi_miR_11143
159 591 4.16
1.0474 0.008867 0.02748 NA
160 687 4.16
0.6025 0.008867 0.02748 NA hsa_miR_302a
161 559 4.14
0.1873 0.009115 0.02805 NA hsa_miR_520e
162 271 4.14
0.2330 0.009167 0.02805 NA hsa_miR_130a
163 71 4.10 0.1119 0.009583 0.02915 NA
hsa_miR_544
164 476 4.09
0.3177 0.009661 0.02921 NA ambi_miR_10766
165 52 4.08 2.3412 0.009844 0.02926 NA
hsa_miR_381
166 523 4.08 0.0852
0.009844 0.02926 NA ambi_miR_7510
167 448 4.07
1.4428 0.009883 0.02926 NA
168 690 4.06
0.4057 0.009974 0.02926 NA hsa_miR_498
169 267 4.06
0.2152 0.009974 0.02926 NA
170 94 4.02 2.1153 0.010391 0.03028 NA
171 24 4.02 1.6504 0.010443 0.03028 NA
172 580 3.97
0.1421 0.011198 0.03228 NA
173 735 3.95
0.3007 0.011289 0.03235 NA hsa_miR_126
174 339 3.92
1.2604 0.011758 0.03350 NA CONTROL_48
175 55 3.88 2.0547 0.012122 0.03434 NA
176 16 3.85 2.1992 0.012591 0.03538 NA
CONTROL_1
177
236 3.84 0.0221 0.012630 0.03538 NA
178
492 3.82 1.8648 0.013034 0.03630 NA
179
587 3.81 1.3354 0.013346 0.03696 NA
180 66 3.79 1.4291 0.013490 0.03715 NA
ambi_miR_7055
181 82 3.78 0.2230 0.013750 0.03766 NA
CONTROL_1
182 73 3.77 2.2194 0.013932 0.03792 NA
hsa_miR_373_AS
183 745 3.76
0.1290 0.013997 0.03792 NA CONTROL_31
184 544 3.74
0.2357 0.014375 0.03873 NA hsa_miR_224
185 257 3.68
1.5474 0.015208 0.04068 NA ambi_miR_13237
186 34 3.68 2.3934 0.015260 0.04068 NA
hsa_miR_330
187 181 3.65
0.2490 0.015781 0.04184 NA hsa_miR_516_5p
188 353 3.63
0.7662 0.016120 0.04251 NA hsa_miR_133b
189 46 3.59 0.1574 0.016771 0.04399 NA
190 371 3.58
0.7029 0.016940 0.04420 NA hsa_miR_19a
191 706 3.55
1.3220 0.017565 0.04536 NA
192 328 3.54
0.1856 0.017565 0.04536 NA hsa_miR_496
193 249 3.49
0.5027 0.018464 0.04725 NA
194 590 3.49 0.1256
0.018490 0.04725 NA hsa_miR_30b
195 647 3.48
0.2040 0.018828 0.04763 NA hsa_miR_539
196 13 3.47 2.4226 0.018984 0.04763 NA
CONTROL_1
197 70 3.47 1.0676 0.018984 0.04763 NA
198 216 3.47
0.0618 0.019023 0.04763 NA
199 542 3.46
0.4283 0.019232 0.04780 NA hsa_miR_182_AS
200 743 3.45
0.6468 0.019284 0.04780 NA ambi_miR_7912
201 226 3.43
0.2089 0.019831 0.04870 NA hsa_miR_31
202 169 3.43
0.6770 0.019844 0.04870 NA CONTROL_1
203 32 3.41 0.4598 0.020456 0.04996 NA
hsa_miR_486
The miRNA lists for this part (SAM) of the project are not the same as for the previous part (significant genes). This suggests that the miRNA that distinguish between ARMS and ERMS or between PRMS and GIST are not the same miRNA that can be used to distinguish between tumor and normal tissue and they are not the same as those miRNA that are the most significant in distinguishing that the 7 types of tissue that have at least 2 samples are not all the same tissue type.
The differences in the methods used in this section and the previous section are that in limma is a linear analysis of microarray data using information from the entire experiment to help estimate standard errors. SAM uses the microarray data permuting the names of the arrays to simulate what a null distribution of data might look like under the same experimental conditions as the microarrays were put under in the real comparison.
I trust the SAM method more than limma for my data, because as we saw in the section on interesting problems with data, there are some very bad arrays (such as ones that do not have any red on them, or others that are completely red on the background too) that I am not confident that normalization will fix entirely. By permuting the names of the arrays that seems as though it would provide a better idea of which miRNA are significant relative to the rest of the data than when using limma which uses a standard error from all the microarray data together.
I think it is safer to trust cutoffs in which there are fewer comparison groups. The normal versus tumor test is easier to trust the cutoffs because the miRNA are just testing the comparison between those two groups. When comparing all of the types of tissue, a miRNA might be really significant if it just distinguishes between two types of tissue really well because in this multiple comparison test, we are only testing if they are all the same versus the tissue types are not all the same.
To me it makes the most sense to trust the FDR (as opposed to FWER or just the significance level) because then it seems like you have more control over how many miRNA you find as significant when they are not supposed to be.
This website was designed by Austen Head.
Email: austen
[dot] head [at]