|
Matthew
H. Sazinsky |
|
|
HOME | TEACHING | RESEARCH | EDUCATION | PUBLICATIONS |
Welcome to our
research page
Chemistry Department, Seaver North, Room 212 Laboratory phone: (909) 607-7959 The Sazinsky
Research Group. Matt Sazinsky, • Click here to see a full list of
undergraduate thesis students •
Click here to see a full list of papers, attended meetings, and abstracts
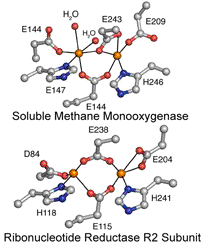
Diiron Enzymes
Carboxylate-bridged
diiron proteins are found in almost all organisms and participate in a variety
of essential biochemical functions, including hydrocarbon and fatty acid
hydroxylation, tyrosyl radical generation, oxidative stress protection, O2
transport and sensing, NO reduction, iron storage, fatty acid
desaturation, and ubiquinol oxidation in mitochondrial membranes. Some of these
proteins, like ribonucleotide reductase (RNR) and soluble methane monooxygenase
(sMMO), have received significant attention because of their biomedical,
industrial, and environmental importance.
The diversity of these powerful O2-utilizing dinuclear active
sites rivals, if not surpasses, that of heme proteins, but diiron enzymes are
found less frequently in nature. Although most diiron proteins share several
structural and mechanistic features, such as strikingly similar dinuclear iron
units that react with O2 and traverse peroxo and/or superoxo
intermediates, it has been particularly challenging to reveal how the protein
scaffold around the metal center governs reactivity. My laboratory aims to investigate the structure/function
relationships responsible for the chemistry and tuning of dinuclear iron active
sites by 1) focusing on proteins that carry out novel reactions using unique
metal coordination spheres and by 2) re-engineering well characterized systems
to perform new functions.
Bacterial
Biofilms
Mature bacterial cells can exist in
two states, as free-floating planktonic cells or as densely packed biofilms on
the surfaces of biological and abiotic materials. In their planktonic form,
pathogenic bacteria species like Streptococcus pneumoniae, Staphalococcus aureus, Salmonella enterica and Pseudomonas aeruginosa are susceptible to antimicrobial
agents. As biofilms, however, these
bacteria are highly resistant to antimicrobials owing to a dense matrix of
extracellular polysaccharides, proteins, and DNA known collectively as the
extracellular polymeric substance (EPS).
Although the general organization and function of the EPS matrix is not
known, it is proposed to promote adhesion between cells and host surfaces and
offer protection from hostile extracellular conditions. Because of the seemingly impenetratable
nature of these films, chronic infections can result, such as in the
respiratory and gastrointestinal tracts of patients with exposure to
opportunistic pathogenic organisms. An understanding of biofilm development,
composition and organization is essential for developing therapies aimed at
disrupting their formation.
Most investigations into bacterial
biofilms have focused on identifying the genetic, molecular, and physiological
determinants of initiation and development. Genome-based microarray analysis and transposon mutagenesis
have identified several intriguing protein targets, but, a universal set of
proteins responsible for this process have not been easy to identify since
different bacterial species do not always use similar machinery for biofilm
formation. Structural and biochemical analysis of biofilm related proteins in
well characterized model organisms will provide a clearer picture of the EPS
composition as well as new avenues to combat chronic infections in a variety of
pathogenic bacteria.
Funding
Camille
and Henry Dreyfus Faculty Start-up Award (2007)
Current Undergraduate Research Students
![]()
