|
HOME | DESIGN | MICROARRAY SAMPLES | NORMALIZATION | SIGNIFICANCE TESTING | SAM | H. CLUSTERING | PAM CLUSTERING | PAM CLASSIFICATION | CONCLUSIONS |

This web page was produced as an assignment for a course on Statistical
Analysis of Microarray Data at
PAM Clustering:
Because of the small size of the dataset, I was able to perform PAM
clustering on the entire miRNA library (768 genes total).
There are two k-values we are interested in biologically: k=2 (healthy
versus tumor tissue) and k=9 (because there are 7 tumor types and 2 healthy
tissue types). However, we will compute PAM clusters for several k-values
to compare the results.
Two separate distance metrics were used - Pearson correlation and Euclidean
distance.
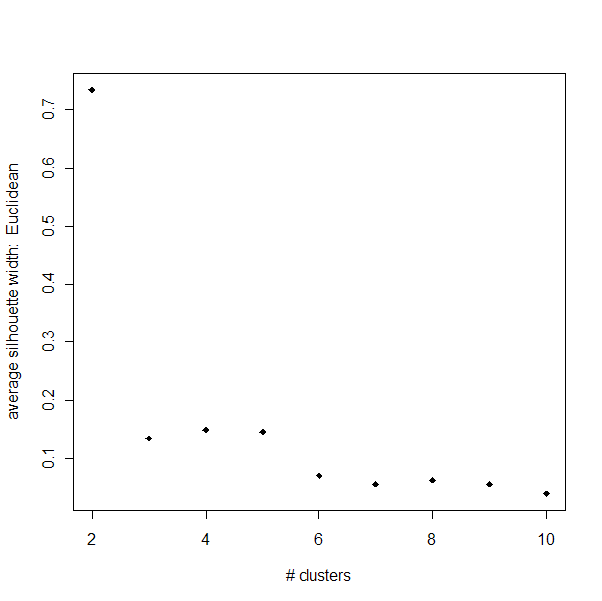
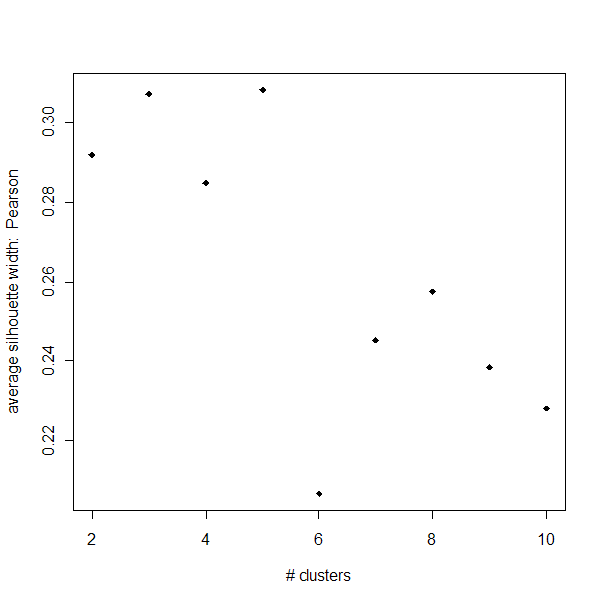
Silhouette Width:
Below is a summary of the Silhouette widths for PAM clusterings
at various k-values using distances based on Pearson correlation as well as
Euclidean distance. Note that silhouette widths close to 1 are desired.
Also note that for neither distance method does k=9 look like the optimal
number of clusters.
|
|
|
There are a couple of interesting things to point out. Our strongest
clusters are with Euclidean distances and k=2. However,
for k>2, the Euclidean clusters peform very
weakly. The Pearson clusters have consistantly
higher silhouette widths than the Euclidean ones for k>2. The Pearson
clusters also have a more interesting pattern - for whatever reason, at k=6,
the silhouette width drops drastically but then returns to stronger values
instantly. These results could indicate that k=9 clusters aren't ideal,
perhaps because some of the tumor types are extremely similar in expression
profiles.
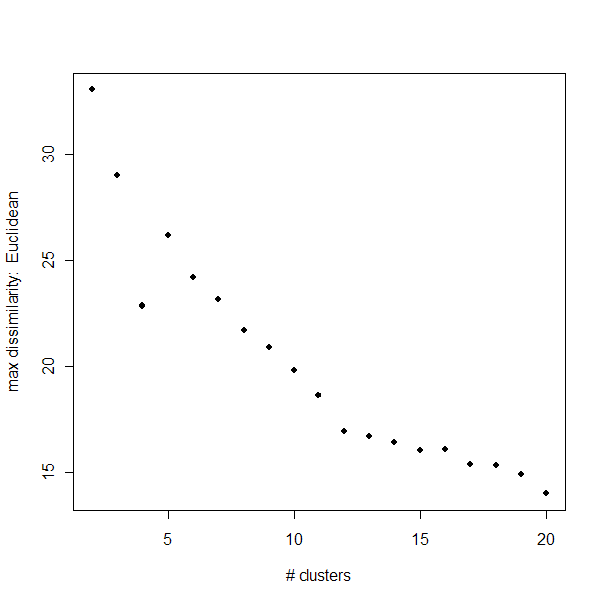
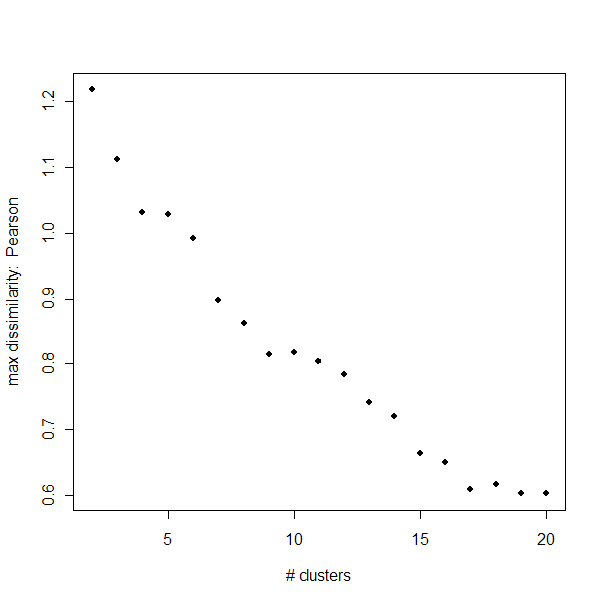
Maximum Dissimilarity:
The maximum dissimilarity is a measure of the maximum distance between
two members of one group. It will continue to decrease until k=n (number
of miRNAs), but it does level off significantly when
the clustering is saturated.
|
|
|
There is a clear
"elbow" to the Euclidean distance dissimilarities around k=11.
For the Pearson dissimilarities, the elbow is not as clear, but could
occur as late as k=17.
Tables
Comparing the clusters of the Euclidean versus Pearson
can add insight into which clustering method is more appropriate. Below
is a comparison of the Euclidean versus Pearson clustering for k=9. The
rows are Pearson clusters, and the columns are Euclidean clusters.
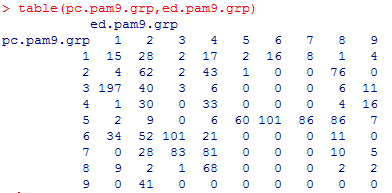
The Pearson cluster #1, and perhaps #2, are evenly
spread out between the 9 Euclidean clusters, but after that the Pearson
clusters seem to correlate to certain subsets of the Euclidean clusters.
The Euclidean clusters seem to be spread out among the Pearson clusters
more consistently, with only #5, #6, #7 and perhaps #3 showing any specificity.
Discussion
It's difficult to determine whether or not the
clusters we have are in fact legitimate. I give much more weight to the
Pearson correlation distances than the Euclidean distances for this analysis.
Thus, the low silhouette widths are discouraging for k=9. However,
for 1<k<5, the silhouette widths are somewhat acceptable, so it might be
worth investigating whether some of the tumor types have very similar
expression profiles (and thus cluster together). I also want to
investigate the two clusters found using the Euclidean distance method to see
if they correlate to tumor versus healthy tissues.
The authors used hierarchical clustering methods, so this PAM analysis would be
completely new.